

Similar to other types of cancer, clonal expansion of abnormal cells is a hallmark of ATL. Our Joint Study on Predisposing Factors of ATL Development (JSPFAD) group examined ATL risk factors and demonstrated that a proviral load (PVL i.e., the percentage of infected peripheral blood mononuclear cells) of >4% is one of the risk factors for progression to ATL however, PVL alone cannot predict development of the disease. Currently, there is no clear determinant to distinguish between individuals who will remain ACs and those who will develop ATL. Whereas the majority of HTLV-1–infected individuals remain asymptomatic carriers (ACs) throughout their lifetime, ~5% of them develop ATL after a long period of clinical latency. HTLV-1 mainly survives in vivo by persistent clonal proliferation of infected cells. HTLV-1 infection and integration of provirus into the host genome are intrinsic and inevitable early events for ATL development. Among the different types of cancer, ATL is a remarkably unique neoplasm in that it is directly caused by infection with human T-cell leukemia virus type-1 (HTLV-1), which is mainly transmitted via breastfeeding. ATL develops through a multistep leukemogenic process, the nature of which remains elusive.

Consequently, focus has shifted toward devising mathematical/computational models for simplification and extraction of fundamental meaning from the complex biological processes of cancer.Īdult T-cell leukemia (ATL) is a life-threatening malignancy that manifests with very poor prognosis. Experimental data alone are not generally sufficient enough to address the complex problem of cancer.
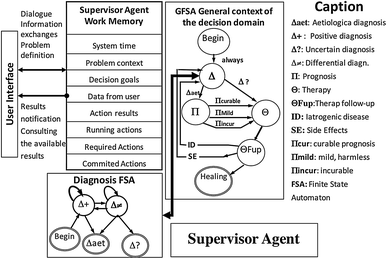
However, the intricate nature of clonal expansion and evolution in cancer makes it difficult to convert the experimental and clinical data into medical practices. The vast amounts of invaluable data generated by NGS have surpassed analysis and interpretation capacity. In recent years, the use of next-generation sequencing (NGS) technologies for the investigation of tumor genomes has generated increasing evidence that most neoplasms grow as a clonally expanded cell population. Since Nowell first proposed the clonal evolution theory of neoplasia in 1976, a broad range of studies have provided support for this model. Although cancer is a diverse and multifactorial disorder with differing origins and degrees of malignancy, clonal expansion and the presence of Darwinian or natural selection are generally accepted as common features. This kind of modeling provides a basic understanding as well as a unique perspective for clarifying the mechanisms of clonal expansion in ATL.Ĭancer is a complex disease of the genome that behaves as a clonal evolutionary process in populations of cells. Our data suggest that combining experimental data (absolute size of clones) with DFA can describe the clonality status of patients. Through the developed model, we have translated biological data of clonal expansion into the formal language of mathematics and represented the observed clonality data with DFA. We propose an empirical formal model based on deterministic finite state automata (DFA) analysis of real clinical samples to illustrate patterns of clonal expansion. We categorized clones into four size groups, “very small”, “small”, “big”, and “very big”, based on the patterns of clonal growth and observed clone sizes. We analyzed clinical samples from HTLV-1–infected individuals with a broad range of proviral loads using a high-throughput methodology that enables isolation of HTLV-1 integration sites and accurate measurement of the size of infected clones. MethodsĪs a comprehensively interdisciplinary project, this study combined three main aspects: wet laboratory experiments, in silico analysis and empirical modeling. Combining computational/mathematical modeling with experimental and clinical data of integration site–based clonality analysis derived from next generation sequencing technologies provides an appropriate strategy to achieve a better understanding of ATL development. However, because of the complex nature of clonal expansion, the underlying mechanisms have yet to be clarified. Therefore, monitoring clonal expansion of HTLV-1–infected cells via isolation of integration sites assists in analyzing infected individuals from early infection to the final stage of ATL development. Infection with human T-cell leukemia virus type-1 (HTLV-1) is the direct cause of ATL onset, and integration of HTLV-1 into the human genome is essential for clonal expansion of leukemic cells. Clonal expansion of leukemic cells leads to onset of adult T-cell leukemia (ATL), an aggressive lymphoid malignancy with a very poor prognosis.


 0 kommentar(er)
0 kommentar(er)
